Here, CCSI (Chromatin Chromatin Space Interaction) database presents 3,017,962 chromatin interaction pairs with annotation of genes, enhancers and SNPs in many cell lines of human, mouse and yeast. These data were obtained by means of 3C, 4C, 5C, ChIA-PET and Hi-C technology in a cell's natural state, nearly all of which detected the three-dimensional architecture of chromosome by coupling ligation in close spatial proximity followed by high-throughput sequencing. So transcriptional regulatory mechanism in disease pathogenesis associated with spatial interactions among genes, enhancers and SNPs could be explored on the base of it.
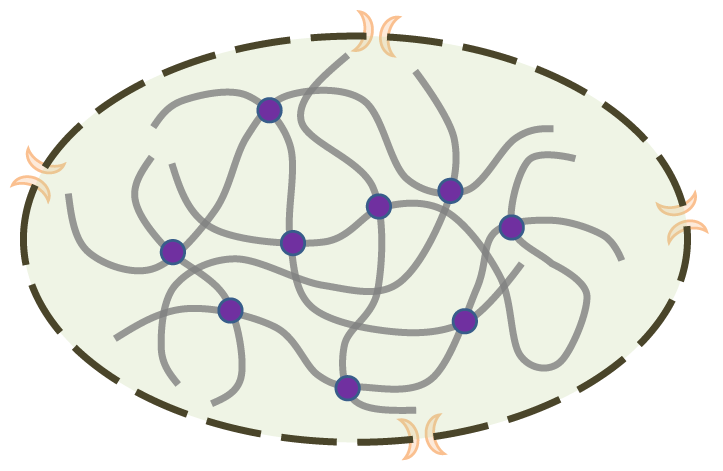
Chromatin folds and contacts with each other in the cell nucleus, which would create opportunity for functional relationship between them. As the picture above describes, the dashed circle with two orange crescents standing for nuclear pore complex is the nucleus membrane. The thick grey lines are chromatin and the purple circles stand for proteins that link chromatin together.
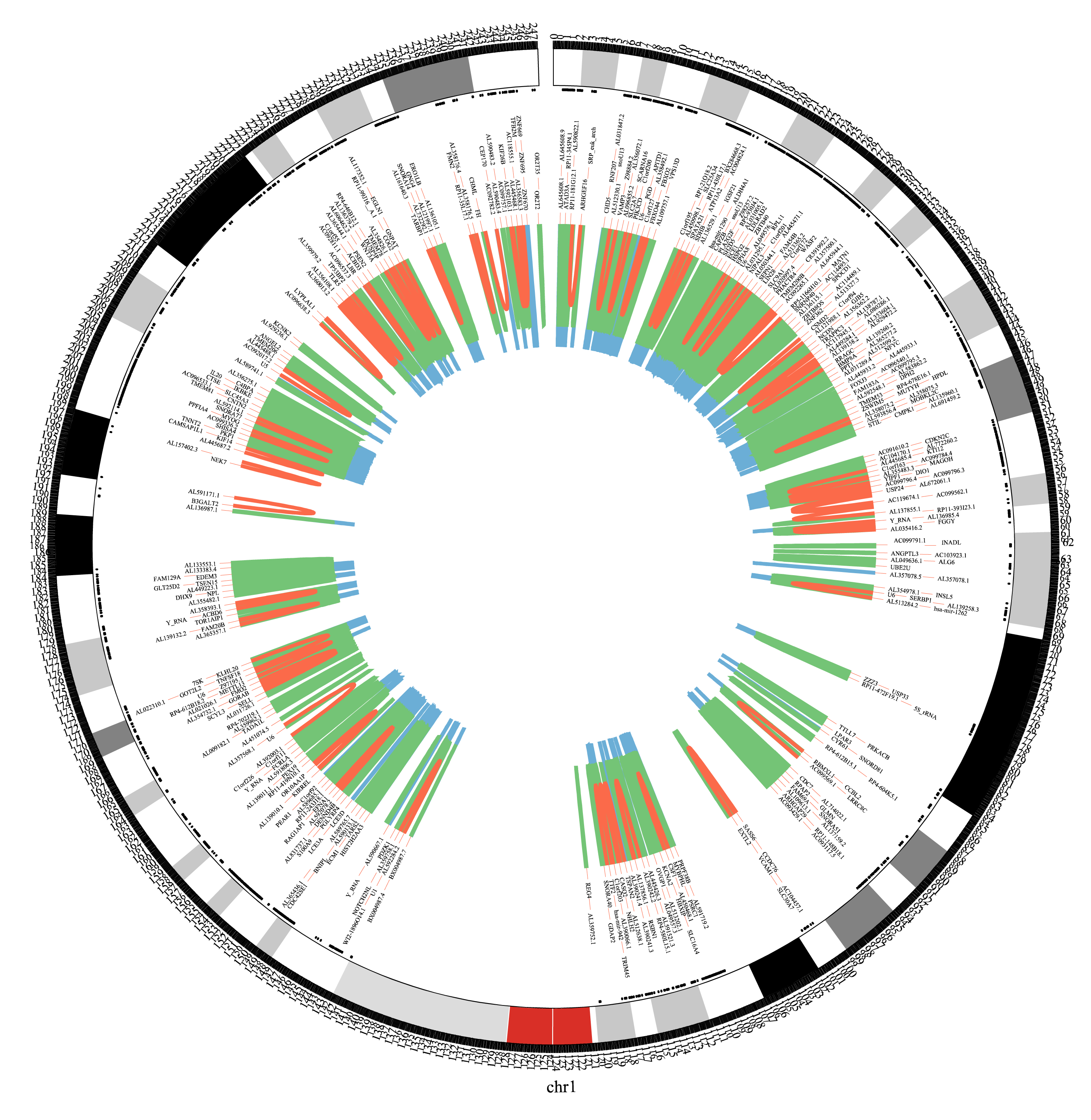
Circos figure is used to describe the promoter-promoter interactions of Chr1 on the basis of a Hi-C dataset. The red lines stand for long-range interactions (distance between two fragments > 500kb), while the blue lines for short-range (distance < 50kb) and the green lines for middle-range (distance spanning 50kb-500kb). The black texts are the gene names of corresponding loci.
Stem cell and functional genomics laboratory at Sun Yat-sen University. If you have some suggestions on the CCSI database, please contact E-mail: xiexw3@mail2.sysu.edu.cn
Tips: For better browse experience, Chrome and Firefox are strongly recommanded.
Citation: Xie, X.W., Ma, W.B., Zhou, S.Y., et al. (2016) CCSI: a database providing chromatin-chromatin spatial interaction information. Database-Oxford