PackPPI
A deep learning framework for side-chain packing and mutation effect prediction.
Deep learning methods have played a crucial role in side-chain packing and mutation effect prediction (ΔΔG) for protein complexes. Although these two tasks are inherently closely related, they are typically treated separately in practice.
To fill this gap, we propose a novel integrated framework called PackPPI, aimed at improving the accuracy and efficiency of side-chain packing and mutation effect prediction for protein complexes. The framework comprises three functional modules:

PackPPI-MSC
Side-chain packing
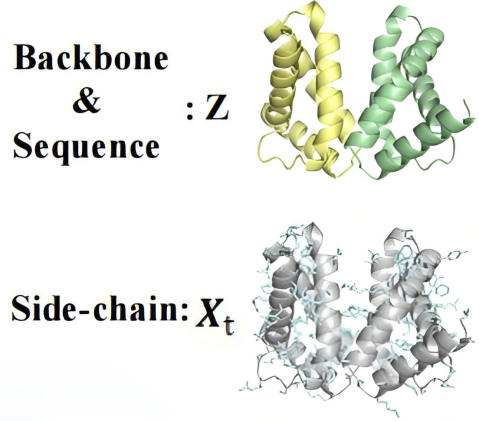
In the PackPPI-MSC module, for each protein complex used in pre-training, a joint noise process is defined on the four torsion angles of side chains. A conditional encoding network then learns the denoising process and protein structure representations.
PackPPI-PROX
Proximal optimization
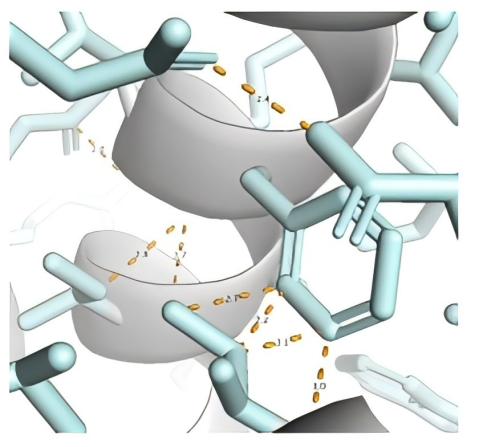
In the PackPPI-Prox module, it uses use a proximal gradient descent method to avoid atomic collisions during sampling, resulting in more reliable side-chain predictions.
PackPPI-AP
Affinity prediction
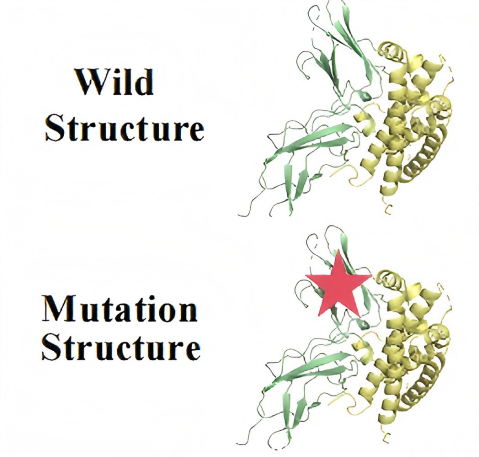
In the PackPPI-AP module, leveraging the encoder layers pretrained in PackPPI-MSC, it first extract pre-trained representations of the structures from each sample in the mutation dataset for both WT and corresponding mutant complex. Subsequently, the mutation encoder is utilized to further extract representation differences caused by mutations. Finally, a multi-layer perceptron serves as a decoder to predict ∆∆G from the combined representations.